Phyleauxgenetics Lab
Phylogenetics and Molecular Evolution
The Lab of Jeremy M. Brown @ Louisiana State University
Phylogenetics and Molecular Evolution
The Lab of Jeremy M. Brown @ Louisiana State University

Undergraduate Researcher

Undergraduate Web Developer
More about the themes of our research
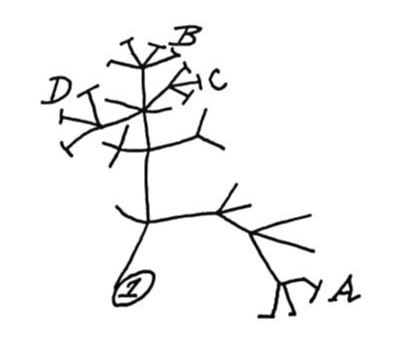
A central tenet of modern biology
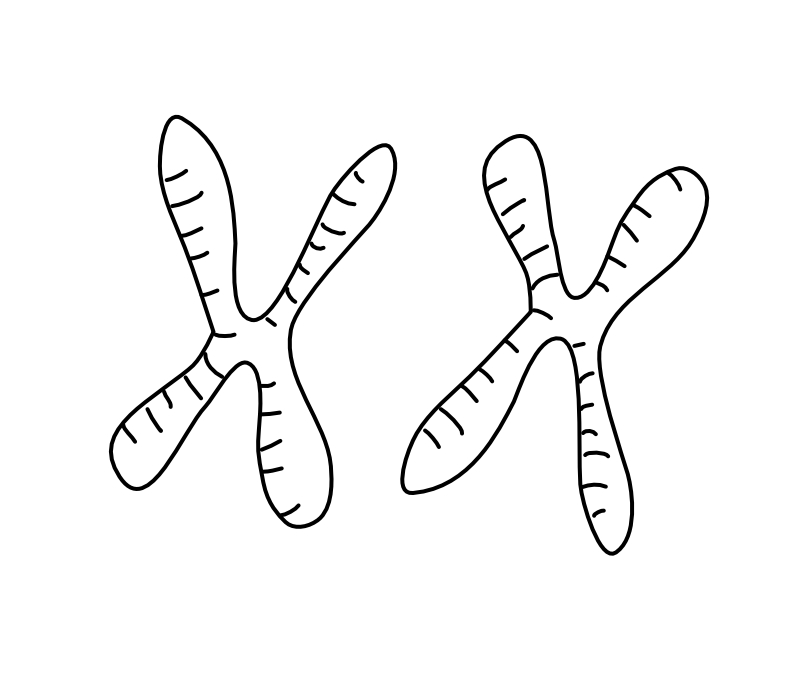
Evolution across the genome
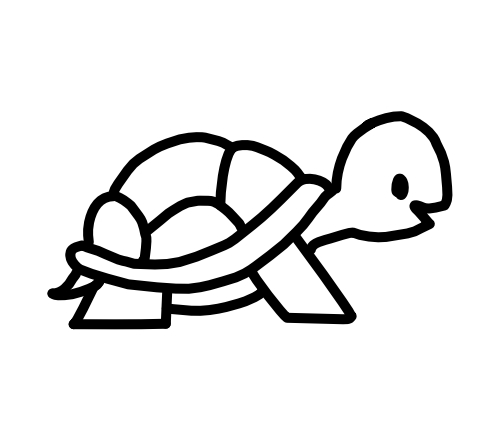
From viruses to turtles
Modeling molecular evolution

P3: Phylogenetic Posterior Prediction
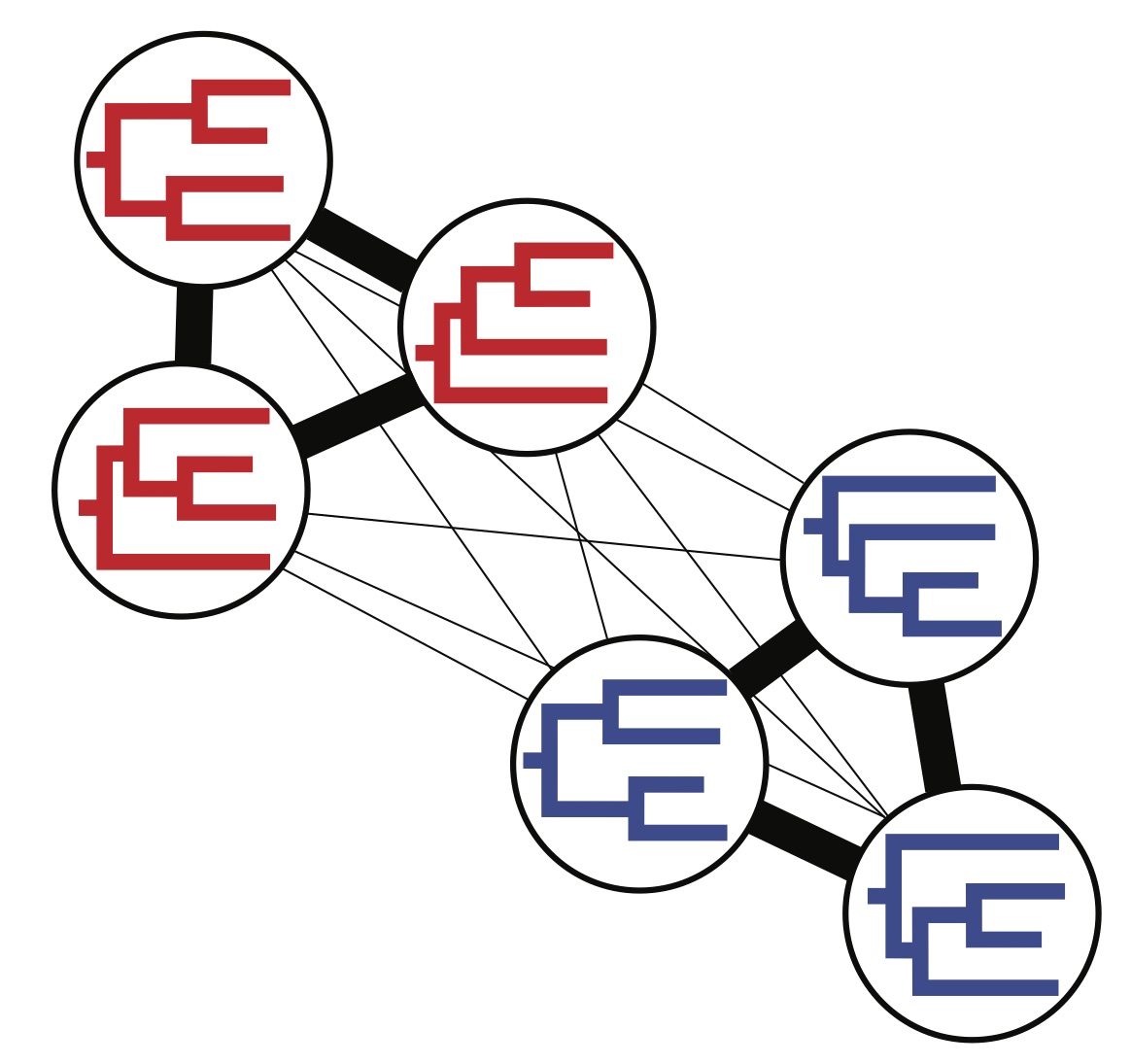
Explore and analyze sets of trees

Analysis of Model Adequacy
Toups, B., T. McGowan, J.M. Brown, K.A. Gallivan, and J.C. Wilgenbusch. 2020. CloudForest: an integrated and dynamic phylogenomic toolset for the modern age. In Review.
Toups, B., and J.M. Brown. 2020. Evolution in the courtroom: Using phylogenetics to investigate legal claims of HIV transmission. In Review.
Hime, P.M., A.R. Lemmon, E.C.M. Lemmon, E. Scott-Prendini, J.M. Brown, R.C. Thomson, J.D. Kratovil, B.P. Noonan, R.A. Pyron, P.L.V. Peloso, M.L. Kortyna, J.S. Keogh, S.C. Donnellan, R.L. Mueller, C.J. Raxworthy, K. Kunte, S. Ron, S. Das, N. Gaitonde, D.M. Green, J. Labisko, J. Che, and D.W. Weisrock. 2020. Phylogenomics reveals ancient gene tree discordance in the amphibian tree of life. In Press. Systematic Biology
Thaljeh, L.F.*, J.A. Rothschild, M. Naderi, L.M. Coghill, J.M. Brown, M. Brylinski. 2019. Hinge region in DNA packaging terminase pUL15 of herpes simplex virus as a potential allosteric target for antiviral drugs. Biomolecules. 9:603.
Rider, P.J.F., L.M. Coghill, M. Naderi, M. Brylinski, J.M. Brown, and K.G. Kousoulas. 2019. Phylogenetics and structural modeling of herpes simplex virus glycoprotein K (gK) identify functionally important domains and residues critical for alphaherpesvirus infection. Scientific Reports. 9:14625.
Brown, J.M. and R.C. Thomson. 2018. Evaluating model performance in evolutionary biology. Annual Review of Ecology, Evolution, and Systematics. 49:95-114.
Richards, E.J., J.M. Brown, A.J. Barley, R.A. Chong, and R.C. Thomson. 2018. Variation across mitochondrial gene trees provides evidence for systematic error: how much gene tree variation is biological? Systematic Biology. 67:847-860.
Brown, J.M. and R.C. Thomson. 2018. The behavior of Metropolis-coupled Markov chains when sampling rugged phylogenetic distributions. Systematic Biology. 67:729-734.
Höhna, S., L.M. Coghill, G.G. Mount, R.C. Thomson, and J.M. Brown. 2018. P3: Phylogenetic Posterior Prediction in RevBayes. Molecular Biology and Evolution. 35:1028-1034.
Spinks, P.Q., E. McCarteny-Melstad, T.J. Near, G.G. Mount, and H.B Shaffer. 2017. Phylogenomic analyses of 539 highly informative loci dates a fully resolved time tree for the major clades of living turtles (Testudines). Molecular Phylogenetics and Evolution. 115:7-15.
Barley, A.J., J.M. Brown, and R.C. Thomson. 2017. Impact of model violations on the inference of species boundaries under the multispecies coalescent. Systematic Biology. 67:269-284.
Brown, J.M. and R.C. Thomson. 2017. Bayes factors unmask highly variable information content, bias, and extreme influence in phylogenomic analyses. Systematic Biology. 66:517-530.
Fujimoto, K., L.M. Coghill, C.A. Weier, L.-Y. Hwang, J.Y. Kim, J.A. Schneider, M.L. Metzker, and J.M. Brown. 2017. Lack of support for socially connected HIV-1 transmission among young adult black MSM. AIDS Research and Human Retroviruses. 33:935-940.
Huang, W., G. Zhou, M. Marchand, J. Ash, D. Morris, P. Van Dooren, J.M. Brown, K.A. Gallivan, and J.C. Wilgenbusch. 2016. TreeScaper: visualizing and extracting phylogenetic signal from sets of trees. Molecular Biology and Evolution. 33:3314-3316.
Engebretsen, K.N., L.N. Barrow, E.N. Rittmeyer, J.M. Brown+, and E.M. Lemmon+. 2016. Quantifying the spatio-temporal dynamics in a chorus frog hybrid zone (Pseudacris) over 30 years. Ecology and Evolution. 6:5013-5031.
+ Co-Senior Authors
Andersen, J.J., B.J. Nelson, and J.M. Brown. 2016. EmpPrior: Using empirical data to inform branch-length priors for Bayesian phylogenetics. BMC Bioinformatics. 17:253.
McCarteny-Melstad, E., G.G. Mount, and H.B. Shaffer. 2016. Exon capture optimization in amphibians with large genomes. Molecular Ecology Resources. 16:1084-1094.
Doyle, V.P., R.E. Young, G.J.P. Naylor, and J.M. Brown. 2015. Can we identify genes with increased phylogenetic reliability? Systematic Biology. 64:824-837.

Nelson, B.J., J.J. Andersen, and J.M. Brown. 2015. Deflating trees: Improving Bayesian branch-length estimates using informed priors. Systematic Biology. 64:441-447.
Couturier, J., D.J. Luke, N. Agarwal, J.W. Suliburk, J.M. Brown, X. Yu, C. Nguyen, D. Iyer, C.A. Kozinetz, P.A. Overbeek, M.L. Metzker, A. Balasubramanyam, and D.E. Lewis. 2015. Human adipose tissue as a reservoir for HIV and activated memory CD4 T cells. AIDS. 29:667-674.
Brown, J.M. 2014. Predictive approaches to assessing the fit of evolutionary models. Systematic Biology. 63: 289-292.
Brown, J.M. 2014. Detection of implausible phylogenetic inferences using posterior predictive assessment of model fit. Systematic Biology. 63: 334-348.
Doyle, V.P., J.J. Andersen, B.J. Nelson, M.L. Metzker, and J.M. Brown. 2014. Untangling the influences of unmodeled evolutionary processes on phylogenetic signal in a forensically important HIV-1 transmission cluster. Molecular Phylogenetics and Evolution. 75:126-137.
Reid, N.M., S.M. Hird, J.M. Brown, T.A. Pelletier, J.D. McVay, J.D. Satler, and B.C. Carstens. 2014. Poor fit to the multispecies coalescent is widely detectable in empirical data. Systematic Biology. 63:322-333.
* Noah Reid awarded Society of Systematic Biologist's Publisher's Award for Best Student Paper of 2014
Spinks, P.Q., R.C. Thomson, G.B. Pauly, C.E. Newman, G.G. Mount, and H.B. Shaffer. 2013. Misleading phylogenetic inferences based on single-exemplar sampling in the turtle genus Pseudemys. Molecular Phylogenetics and Evolution. 68:269-281.
Fong, J.J., J.M. Brown, M.K. Fujita, and B. Boussau. 2012. A phylogenomic approach to vertebrate phylogeny supports a turtle-archosaur affinity and a possible paraphyletic Lissamphibia. PLoS ONE 7:e48990.
Boussau, B., J.M. Brown, and M.K. Fujita. 2011. Nonadaptive evolution of mitochondrial genome size. Evolution. 65:2706-2711.
Leitner, T., M.L. Metzker, D. Zwickl, J.M. Brown, A.M. Geretti, A. Vandamme, J. Albert, and D.M. Hillis. 2011. Forensic HIV phylogenetics. Nature. 473:284 [Correspondence]
Brown, J.M., K. Savidge*, and E.J.B. McTavish. 2010. DIM SUM: Demography and Individual Migration Simulated Using a Markov chain. Molecular Ecology Resources. 11:358-363.
Scaduto, D.I., J.M. Brown, W.C. Haaland, D.J. Zwickl, D.M. Hillis, and M.L. Metzker. 2010. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proceedings of the National Academy of Sciences USA. 107:21242-21247.
Brown, J.M., S.M. Hedtke, A.R. Lemmon, and E.M. Lemmon. 2009. When trees grow too long: investigating the causes of highly inaccurate Bayesian branch-length estimates. Systematic Biology. 59:145-161.
Brown, J.M. and R. ElDabaje*. 2009. PuMA: Bayesian analysis of partitioned (and unpartitioned) model adequacy. Bioinformatics. 25:537-538.
Lemmon, A. R., J.M. Brown, K. Stanger-Hall, and E.M. Lemmon. 2009. The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Systematic Biology. 58:130-145.
Rabeling, C., J.M. Brown, and M. Verhaagh. 2008. Newly discovered sister lineage sheds light on early ant evolution. Proceedings of the National Academy of Sciences USA. 105:14913-14917.
Brown, J.M. and A.R. Lemmon. 2007. The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Systematic Biology. 56:643-655.
Brown, J.M. and G.B. Pauly. 2005. Increased rates of molecular evolution in an equatorial plant clade: an effect of environment or phylogenetic non-independence? Evolution. 59:238-242.
Agrawal, A.F., J.M. Brown, and E.D. Brodie III. 2004. On the social structure of offspring rearing in the burrower bug, Sehirus cinctus (Hemiptera: Cydnidae). Behavioral Ecology and Sociobiology. 57:139-148.
Brown, J.M., A.F. Agrawal, and E.D. Brodie III. 2003. An analysis of single clutch paternity in the burrower bug Sehirus cinctus using microsatellites. Journal of Insect Behavior. 16:731-745.
Agrawal, A.F., E.D. Brodie III, and J. Brown*. 2001. Parent-offspring coadaptation and the dual genetic control of maternal care. Science. 292:1710-1712.
* Undergraduate